ESR2: Evolution of the small intestine microbiome in pigs from birth to post weaning under normal and stress conditions
Background
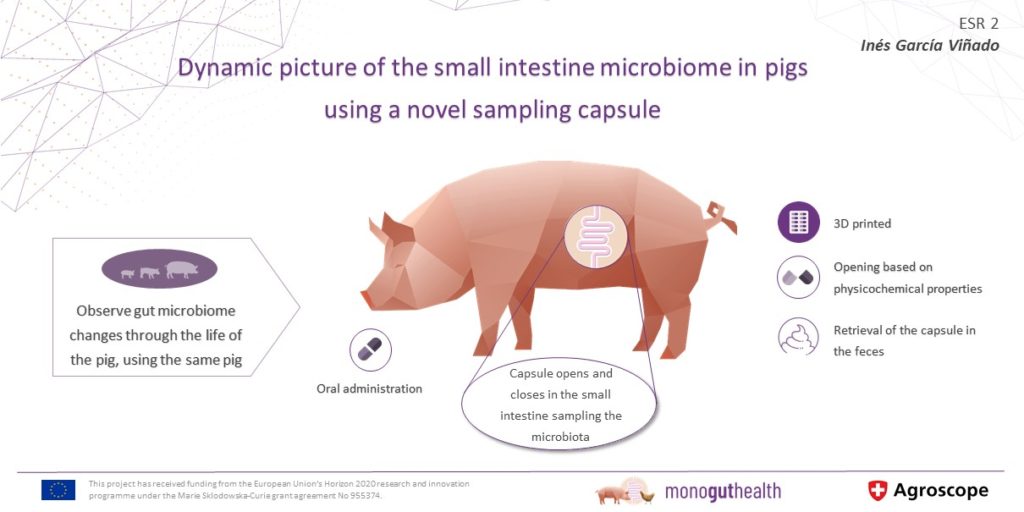
Targeted manipulation of the gut microbiota is increasingly recognized as a mean to improve livestock health. We know that the interpretation of bacterial communities of the small intestineA long tube-like organ that connects the stomach and the large intestine. It helps to digest food co… by studying the rectal microbiomeThe diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome f… from stool samples has its limitations. Thus, getting a more in depth understanding of the precise role of the intestinal microbiomeThe diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome f… in health and disease requires a method to access the ‘true’ (not fecal) intestinal microbiomeThe diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome f….
Objectives
Set up and validate an in vivo sampling tool (e.g. capsule endoscopy) to collect small intestineA long tube-like organ that connects the stomach and the large intestine. It helps to digest food co... content in living pigs.
Determine to what extent the small intestineA long tube-like organ that connects the stomach and the large intestine. It helps to digest food co... microbiomeThe diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome f... changes over time and to assess the temporal stability of an individual pig’s gut microbiomeCollective genetic material of the microbes (for example bacteria, fungi and viruses) that live insi....
Compare previous results with pathophysiologic conditions to evaluate the consequences of an ETECAn important cause of bacterial diarrhea during the post weaning period in the life of a pig. It is ... infection, as a model for pathogens interference.
Methods
In a first step, the new in vivo sampling tool will be optimized and validated.
In subsequent steps, the microbiomeThe diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome f... development through pigs life cycle will be monitored under normal and pathophysiological conditions.
Expected results
Novel in vivo sampling tool for small intestineA long tube-like organ that connects the stomach and the large intestine. It helps to digest food co... content will be validated and standard operating procedure prepared (D2.1).
Extended knowledge on the evolution of phylotypes and/or bacterial species in the small intestineA long tube-like organ that connects the stomach and the large intestine. It helps to digest food co... microbiomeThe diverse consortium of bacteria, archaea, fungi, protozoa, viruses, and their collective genome f... from post weaning to slaughter (D2.4) under physiological and pathophysiological conditions (D3.7).
Planned secondments
- At: UNIBO (7 mo); Bioinformatics for microbiome and metabolome data;
- At: TwentyGreen (1 mo); training in specific microbiological analysis of prebiotics and get inside on how to develop customer information.
Enrolment in Doctoral degree:
ESR2 will be enrolled at the Department of Agricultural and Food Science (DISTAL) of University of Bologna.
Supervisors
Catherine Ollagnier (Agroscope), Giuseppe Bee (Agroscope), Paolo Trevisi (University of Bologna)